Lopsided_Amoeba_Approximation
"Lopsided_Amoeba_Approximation" is a software package for the approximation of amoebas via lopsided amoebas and cyclic resultants. The package consists of "Lopsided_Amoeba_Approximation.lib", a SINGULAR library, for the algebraic operations and two SAGE scripts providing the corresponding graphical outputs.
Version
- Current Version: 0.1.0.0(a)
- Latest Update: 08/30/2016.
Please note that this software is an alpha version. As such it is preliminary and subject to change without notice.
Also note that "Lopsided_Amoeba_Approximation.lib" is not part of the latest SINGULAR distribution yet. It will be submitted to the SINGULAR distributors after it has been tested sufficiently.
Feedback, suggestions, and bug reports are very welcome; please contact me via email.
Developers
- Developer: Timo de Wolff.
- Scientific support & software testing: Laura Felicia Matusevich, Jens Forsgård, Nathan Mehlhop.
Download
- The software package "Lopsided_Amoeba_Approximation": [download].
- All data that was created by "Lopsided_Amoeba_Approximation" and used in the article "Lopsided Approximation of Amoebas": [download].
- A SINGULAR script that was written for the article "Lopsided Approximation of Amoebas" to compare the runtime of the method to compute cyclic resultants developed in this article with the traditional way to compute these objects: [download].
Action Shots
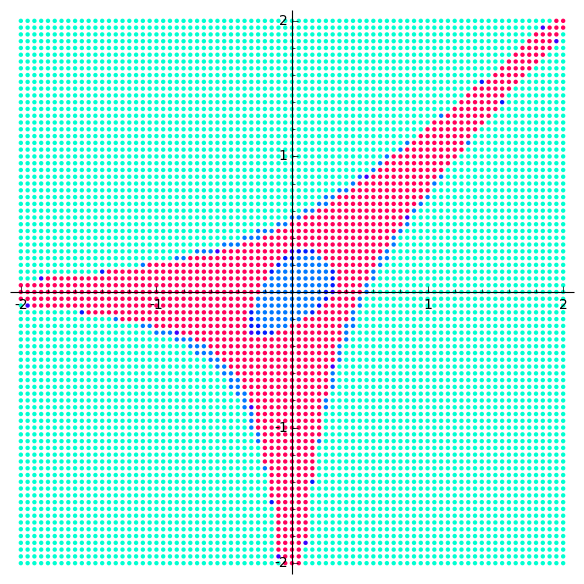
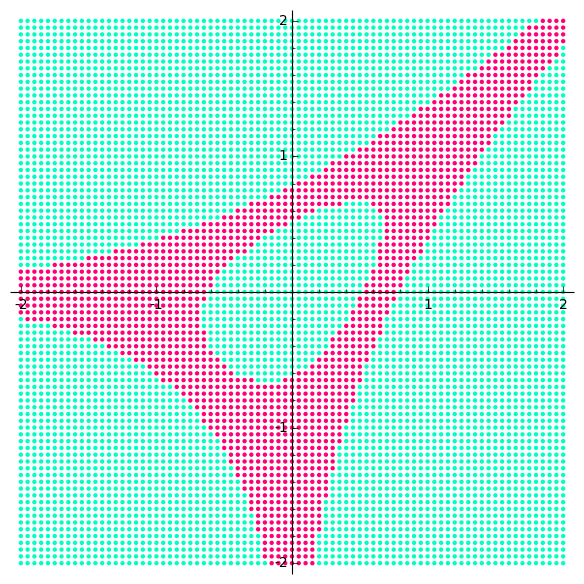
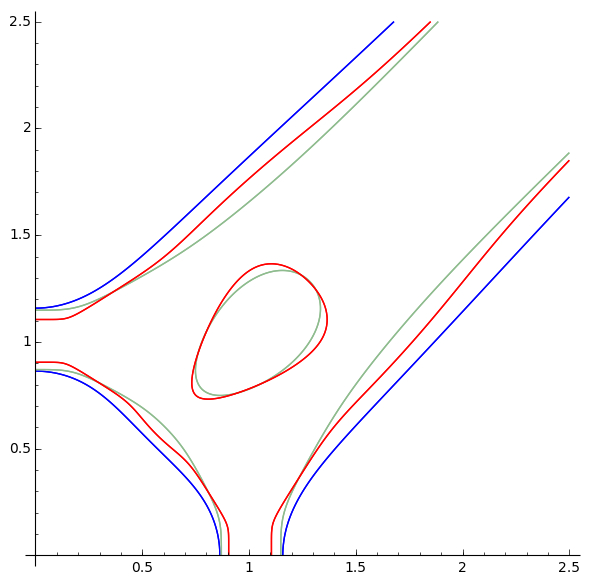
Usage
To set up your machine to use "Lopsided_Amoeba_Approximation", proceed as follows:
- Install SINGULAR on your machine. Instructions for doing so can be found here.
- If you want to use the graphical output of "Lopsided_Amoeba_Approximation", then install SAGE to your machine. Instructions for doing so can be found here.
- You download our software package; save it into "~/YOUR_FOLDER/".
- Start SINGULAR in "~/YOUR_FOLDER/". To load our SINGULAR library execute the command:
> LIB "Lopsided_Amoeba_Approximation.lib"; - You can now use the software.
-
We define a ring R in two variables x,y with imaginary unit I:
> ring R = (0,I),(x,y),lp;
> minpoly = I^2 + 1;
- Define a polynomial:
> poly f = x3 + y3 + 2*x*y + 1; - Compute the corresponding level 3 cyclic resultant:
> poly g = quickcyclicresultant(f,3); - Test whether f is lopsided at the point (1/4,1/4):
> list point = 1/4, 1/4;
> lopsided(f,point); - Generate an 33 x 33 grid on [-2,2]^2 (both with regular and exponential coordinates):
> list testlist = createtestlist(-2,2,-2,2,1/8,1/8); - Test all gridpoints for lopsidedness up to level 3 cyclic resultants of f:
> list results = lopsidedamoeba(f,testlist[2],3);
Note: Evaluating large grids on a high level has a high runtime. Use a divide-and-conquer approach instead. - Save the output in "~/YOUR_FOLDER/amoebaoutputfile.txt":
> saveoutput(testlist[1],results); - Indepently, you can compute the semi-algebraic approximation of the unlog amoeba of f. For plotting your results in SAGE you need to enforce a long output first:
> short = 0;
In this example, we allow cyclic resultants up to level 3:
> list boundaryequations = lopsidedboundary(f,g,3); - Save the output in "~/YOUR_FOLDER/boundaryoutputfile.txt":
> saveboundary(boundaryequations);
-
We define a ring R in two variables x,y with imaginary unit I:
- To display your results start SAGE, load, and execute the scripts "AmoebaLopsidedPlot.sage" and "UnlogAmoebaBoundaryPlot.sage" respectively in SAGE. Make sure that you put in the correct "~/PATH_TO_YOUR_FOLDER/" in the script. The pictures are by default saved as:
- "./AmoebaPlot.png" and
- "./AmoebaBoundaryPlot.png".
License
The Software is published under GNU public license.
Literature
- J. Forsgård, L. Matusevich, N. Mehlhop, T. de Wolff: "Lopsided Approximation of Amoebas",
to appear in Math. Comp.; see ArXiv 1608.08663. - K. Purbhoo: "A Nullstellensatz for Amoebas",
Duke Mathematical Journal, 14 (3), 2008, 408-445; ArXiv Version.
Support
This software was written jointly with the article "Lopsided Approximation of Amoebas". This work was supported by the NSF grant DMS - 1500832 (PI: L. Matusevich).